-Search query
-Search result
Showing 1 - 50 of 346 items for (author: yu & lt)

EMDB-18373: 
cryo-EM structure of apo Clostridioides difficile toxin B
Method: single particle / : Kinsolving J, Bous J, Structural Genomics Consortium (SGC)

EMDB-18374: 
cryo-EM structure complex of Frizzled-7 and Clostridioides difficile toxin B
Method: single particle / : Kinsolving J, Bous J

PDB-8qen: 
cryo-EM structure of apo Clostridioides difficile toxin B
Method: single particle / : Kinsolving J, Bous J, Structural Genomics Consortium (SGC)

PDB-8qeo: 
cryo-EM structure complex of Frizzled-7 and Clostridioides difficile toxin B
Method: single particle / : Kinsolving J, Bous J

EMDB-41816: 
Cryo-EM structure of the RAF1-HSP90-CDC37 complex in the closed state
Method: single particle / : Finci LI, Simanshu DK

EMDB-41817: 
Cryo-EM structure of the HSP90 dimer (NTD-MD) in the semi-open state
Method: single particle / : Finci LI, Simanshu DK

EMDB-41818: 
Cryo-EM structure of the cross-linked HSP90 dimer (NTD-MD) in the semi-open state
Method: single particle / : Finci LI, Simanshu DK

PDB-8u1l: 
Cryo-EM structure of the RAF1-HSP90-CDC37 complex in the closed state
Method: single particle / : Finci LI, Simanshu DK

PDB-8u1m: 
Cryo-EM structure of the HSP90 dimer (NTD-MD) in the semi-open state
Method: single particle / : Finci LI, Simanshu DK

PDB-8u1n: 
Cryo-EM structure of the cross-linked HSP90 dimer (NTD-MD) in the semi-open state
Method: single particle / : Finci LI, Simanshu DK

EMDB-16229: 
Cryo-EM structure of the bacterial replication origin opening basal unwinding system
Method: single particle / : Pelliciari S, Bodet-Lefevre S, Murray H, Ilangovan A

PDB-8btg: 
Cryo-EM structure of the bacterial replication origin opening basal unwinding system
Method: single particle / : Pelliciari S, Bodet-Lefevre S, Murray H, Ilangovan A

EMDB-37631: 
Hepatitis B virus capsid (HBV core protein)
Method: single particle / : Yip RPH, Lai LTF, Lau WCY, Ngo JCK, Kwok DCY

EMDB-37634: 
SR protein kinase 2 bound at 2-fold vertex of Hepatitis B virus capsid
Method: single particle / : Yip RPH, Lai LTF, Kwok DCY, Lau WCY, Ngo JCK

EMDB-38062: 
Hepatitis B virus capsid in complex with SR protein kinase 2
Method: single particle / : Yip RPH, Lai LTF, Kwok DCY, Lau WCY, Ngo JCK

EMDB-41830: 
Lipidated recombinant apolipoprotein E4
Method: single particle / : Strickland MR, Rau M, Summers B, Basore K, Wulf II J, Jiang H, Chen Y, Ulrich JD, Randolph GJ, Zhang R, Fitzpatrick JAJ, Cashikar AG, Holtzman DM

EMDB-41831: 
Gradient-fixed lipidated recombinant apolipoprotein E4
Method: single particle / : Strickland MR, Rau M, Summers B, Basore K, Wulf II J, Jiang H, Chen Y, Ulrich JD, Randolph GJ, Zhang R, Fitzpatrick JAJ, Cashikar AG, Holtzman DM

EMDB-17208: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-17212: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 snoRNA mutant
Method: single particle / : Rajan KS, Yonath A

EMDB-17249: 
CRYO-EM CONSENSUS MAP OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A
EMDB-17250: 
CRYO-EM FOCUSED REFINEMENT MAPS OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-17254: 
CRYO-EM CONSENSUS MAP OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 SKO
Method: single particle / : RAJAN KS, YONATH A
EMDB-17255: 
CRYO-EM FOCUSED REFINEMENT OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 snoRNA mutant
Method: single particle / : Rajan KS, Yonath A

PDB-8ova: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

PDB-8ove: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 snoRNA mutant
Method: single particle / : Rajan KS, Yonath A

EMDB-41075: 
SARS-CoV-2 spike in complex with Fab 71281-33
Method: single particle / : Binshtein E, Crowe JE

EMDB-41076: 
SARS-CoV-2 spike in complex with Fab 71281-33 (2)
Method: single particle / : Binshtein E, Crowe JE

EMDB-29281: 
Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2
Method: single particle / : Li J, Canham SM, Zhang X, Bai X, Feng Y

EMDB-29282: 
Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53
Method: single particle / : Li J, Canham SM, Zhang X, Bai X, Feng Y

PDB-8flk: 
Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2
Method: single particle / : Li J, Canham SM, Zhang X, Bai X, Feng Y

PDB-8flm: 
Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53
Method: single particle / : Li J, Canham SM, Zhang X, Bai X, Feng Y

EMDB-34715: 
Cryo-EM structure of ComA bound to its mature substrate CSP peptide
Method: single particle / : Yu L, Xin X, Min L
EMDB-36882: 
Cryo-EM structure of nucleotide-bound ComA with ZinC ion
Method: single particle / : Yu L, Xin X, Min L, Feng H

EMDB-36936: 
Cryo-EM structure of nucleotide-bound ComA E647Q mutant with Mg2+
Method: single particle / : Yu L, Xin X, Min L

EMDB-34712: 
Cryo-EM structure of nucleotide-bound ComA at outward-facing state with EC gate closed conformation
Method: single particle / : Yu L, Xin X, Min L

EMDB-34713: 
Cryo-EM structure of nucleotide-bound ComA at outward-facing state with EC gate open conformation
Method: single particle / : Yu L, Xin X, Min L

EMDB-34714: 
Cryo-EM structure of nucleotide-bound ComA E647Q mutant
Method: single particle / : Yu L, Xin X, Min L

EMDB-34716: 
Cryo-EM structure of ComC bound ComA C17A at inward-facing state
Method: single particle / : Lin Y, Xin X, Min L

EMDB-40184: 
Structure of the Spizellomyces punctatus Fanzor (SpuFz) in complex with omega RNA and target DNA
Method: single particle / : Xu P, Saito M, Zhang F

PDB-8gkh: 
Structure of the Spizellomyces punctatus Fanzor (SpuFz) in complex with omega RNA and target DNA
Method: single particle / : Xu P, Saito M, Zhang F
EMDB-29248: 
Cryo-EM Structure of PG9RSH DU011 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD
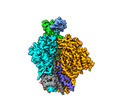
EMDB-29264: 
Cryo-EM Structure of PG9RSH DU025 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD
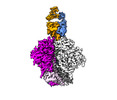
EMDB-29288: 
Cryo-EM Structure of PGT145 DU303 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

PDB-8fk5: 
Cryo-EM Structure of PG9RSH DU011 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

PDB-8fl1: 
Cryo-EM Structure of PG9RSH DU025 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

PDB-8flw: 
Cryo-EM Structure of PGT145 DU303 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

EMDB-34276: 
Cryo-EM structure of CB2-G protein complex
Method: single particle / : Wu LJ, Hua T, Liu ZJ, Li XT, Chang H

EMDB-34277: 
Cryo-EM structure of CP-CB2-G protein complex
Method: single particle / : Wu LJ, Hua T, Liu ZJ, Li XT, Chang H

EMDB-34278: 
Cryo-EM structure of HU-CB2-G protein complex
Method: single particle / : Wu LJ, Hua T, Liu ZJ, Li XT, Chang H

EMDB-34279: 
Cryo-EM structure of LEI-CB2-Gi complex
Method: single particle / : Liu ZJ, Hua T, Li XT, Chang H, Wu LJ
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
